Thank you for pointing out the mistake.
Re-run model select Table
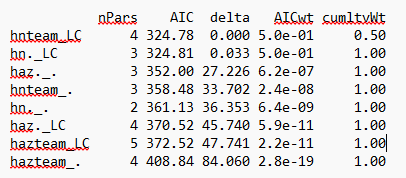
ModSelect
nPars AIC delta AICwt cumltvWt
hnLC_. 3 271.39 0.00 1.0e+00 1.00
hnLC_Team 4 324.78 53.39 2.6e-12 1.00
haz._. 3 352.00 80.61 3.1e-18 1.00
hn._. 2 361.13 89.74 3.3e-20 1.00
hn._Team 3 363.11 91.72 1.2e-20 1.00
haz._LC 4 370.52 99.13 3.0e-22 1.00
hazTeam_LC 5 372.52 101.13 1.1e-22 1.00
hazTeam_. 4 408.84 137.45 1.4e-30 1.00
Call:
distsamp(formula = ~Team ~ Landcover, data = TruncUMF, keyfun = "halfnorm",
output = "density", unitsOut = "ha")
Density:
Estimate SE z P(>|z|)
(Intercept) -2.38 0.128 -18.7 7.86e-78
LandcoverWetland -1.04 0.179 -5.8 6.57e-09
Detection:
Estimate SE z P(>|z|)
(Intercept) 4.502 0.112 40.30 0.000
TeamB 0.195 0.135 1.44 0.149
AIC: 324.777
hazTeam_LC # show model output
Call:
distsamp(formula = ~Team ~ Landcover, data = TruncUMF, keyfun = "hazard",
output = "density", unitsOut = "ha")
Density:
Estimate SE z P(>|z|)
(Intercept) -2.49 0.105 -23.60 3.47e-123
LandcoverWetland -1.07 0.178 -6.01 1.89e-09
Detection:
Estimate SE z P(>|z|)
(Intercept) -11.08 4099 -0.00270 0.998
TeamB -8.88 2892 -0.00307 0.998
Hazard-rate(scale):
Estimate SE z P(>|z|)
-14.4 NaN NaN NaN
AIC: 372.5179
Question 1: The detection and density estimate outputs only show assessments for one category (e.g., Wetland and Team B). How do you get estimate outputs for grassland and team A?
I also tried evaluating the covariate models:
Model selection based on AIC:
K AIC Delta_AIC AICWt Cum.Wt LL
hnLC_. 3 271.39 0.00 1 1 -132.70
hnLC_Team 4 324.78 53.39 0 1 -158.39
haz._. 3 352.00 80.61 0 1 -173.00
hn._. 2 361.13 89.74 0 1 -178.56
hn._Team 3 363.11 91.72 0 1 -178.56
haz._LC 4 370.52 99.13 0 1 -181.26
hazTeam_LC 5 372.52 101.13 0 1 -181.26
hazTeam_. 4 408.84 137.45 0 1 -200.42
Model selection based on AICc:
K AICc Delta_AICc AICcWt Cum.Wt LL
hnLC_. 3 274.39 0.00 1 1 -132.70
hnLC_Team 4 330.49 56.10 0 1 -158.39
haz._. 3 355.00 80.61 0 1 -173.00
hn._. 2 362.46 88.07 0 1 -178.56
hn._Team 3 366.11 91.72 0 1 -178.56
haz._LC 4 376.23 101.84 0 1 -181.26
hazTeam_LC 5 382.52 108.13 0 1 -181.26
hazTeam_. 4 414.55 140.16 0 1 -200.42
Model selection based on QAICc:
(c-hat estimate = 1.679429)
K QAICc Delta_QAICc QAICcWt Cum.Wt Quasi.LL
hnLC_. 4 171.74 0.00 1 1 -79.01
hnLC_Team 5 208.62 36.88 0 1 -94.31
haz._. 4 219.74 48.00 0 1 -103.01
hn._. 3 221.65 49.91 0 1 -106.32
hn._Team 4 226.35 54.62 0 1 -106.32
haz._LC 5 235.86 64.12 0 1 -107.93
hazTeam_LC 6 244.66 72.92 0 1 -107.93
hazTeam_. 5 258.67 86.94 0 1 -119.34
Further problem: I couldn’t get the code right for the summed model weight for team and landcover covariates.
 from your best model
from your best model for density, detectability and ESHW, including confidence intervals
for density, detectability and ESHW, including confidence intervals What conclusion do your draw from your results?
What conclusion do your draw from your results? on your next step, challenge you solved, problem you still have or leap of understanding
on your next step, challenge you solved, problem you still have or leap of understanding to a coursemate
to a coursemate