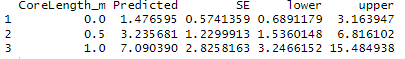Trying to share my results for this assignment:
I worked with the lemur dataset to answer the following questions:
1 – An Increased % of core forest within the transect will decrease the detectability of lemurs.
2 - The less disturbed forest, Anabohazo Forest, will have more density of lemurs as there is less anthropogenic disturbance.
Checked the hypothesis for the half-normal and the hazard detection function.
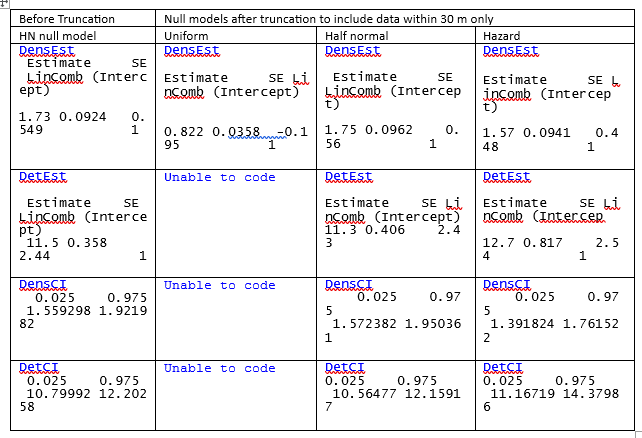
15% of the data fell within 22.5m, however I used 25m to truncate the dataset.
Results:
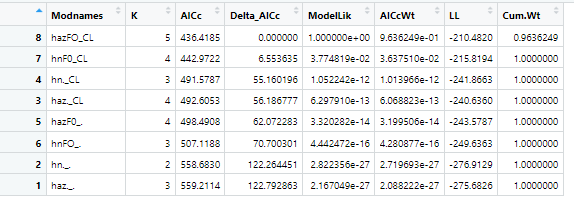
The results showed that the best model that fitted the data was the half normal detection function with Core percent and the Forest as covariates, as seen in the AICc table:
Model selection based on AICc:
K AICc Delta_AICc AICcWt Cum.Wt LL
hnCO_FO 4 474.86 0.00 0.98 0.98 -231.76
hzCO_FO 5 482.67 7.81 0.02 1.00 -233.61
hz._FO 4 506.36 31.51 0.00 1.00 -247.51
hn._FO 3 506.44 31.58 0.00 1.00 -249.30
hnCO_. 3 588.71 113.86 0.00 1.00 -290.43
hzCO_. 4 594.80 119.95 0.00 1.00 -291.73
hz._. 3 609.83 134.97 0.00 1.00 -300.99
hn._. 2 610.40 135.55 0.00 1.00 -302.77
Best fit model:
hnCO_FO
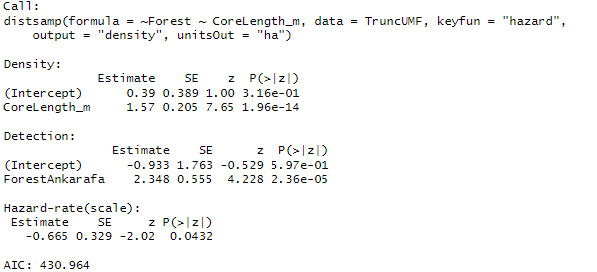
Call:
distsamp(formula = ~CorePercent ~ Forest, data = lem_tru_UMF,
keyfun = "halfnorm", output = "density", unitsOut = "ha")
Density:
Estimate SE z P(>|z|)
(Intercept) 0.028 0.0871 0.321 7.48e-01
ForestAnkarafa 0.997 0.0963 10.355 3.99e-25
Detection:
Estimate SE z P(>|z|)
(Intercept) 1.978 0.0738 26.78 4.91e-158
CorePercent 0.788 0.1565 5.04 4.73e-07
AIC: 471.5221
Density estimates:
Anabohazo: 1.03 lemurs/ha; ranging from 0.87 lemurs/ha to 1.22 lemurs/ha
Ankarafa: 2.8 lemurs/ha; ranging from 2.45 lemurs/ha to 3.17 lemurs/ha
The more disturbed forest, Ankarafa, has rather higher density of lemurs than the less disturb forest.
Detectabiliity estimates:
With 10% of core forest within the transect detectability rate is at 7.82m from the transect.
With a 60% of core forest within the transect detectability rate decreases to 11.6m.
Therefore our Null hyposthesis is true, an increased percentage of core forest within our transect decreases the detectability of the lemurs.
I wanted to go further to asses how good was the model used, however, I am stacked again.
How reliable is our model?
GOF
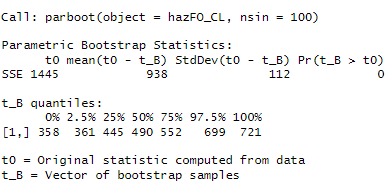
Call: parboot(object = hnCO_FO, nsim = 1000)
Parametric Bootstrap Statistics:
t0 mean(t0 - t_B) StdDev(t0 - t_B) Pr(t_B > t0)
SSE 1436 950 98 0
t_B quantiles:
0% 2.5% 25% 50% 75% 97.5% 100%
[1,] 244 327 414 478 548 690 1026
t0 = Original statistic computed from data
t_B = Vector of bootstrap samples
The p-value is 0 in this case, meaning that our model is a poor fit for this data.
I then calculated the estimated value of ^c to try and improve the model:
cHat estimate:
SSE
2.994418
Seems there is a lack of independence of observations
K QAICc Delta_QAICc QAICcWt Cum.Wt Quasi.LL
hnCO_FO 5 170.25 0.00 0.93 0.93 -77.40
hzCO_FO 6 176.43 6.18 0.04 0.97 -78.01
hn._FO 4 177.84 7.59 0.02 1.00 -83.25
hz._FO 5 180.77 10.52 0.00 1.00 -82.66
hnCO_. 4 205.32 35.07 0.00 1.00 -96.99
hn._. 3 210.07 39.82 0.00 1.00 -101.11
hzCO_. 5 210.31 40.06 0.00 1.00 -97.43
hz._. 4 212.37 42.12 0.00 1.00 -100.52
SumWt_Forest <- importance(models,
c.hat = cHat,
parm = "Forest",
parm.type= "lambda",
second.order = TRUE)
Importance values of 'lam(ForestAnkarafa)':
w+ (models including parameter): 1
w- (models excluding parameter): 0
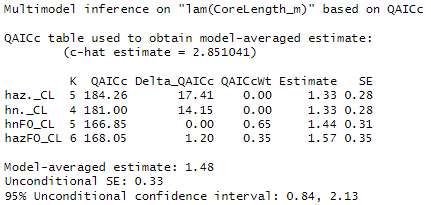
And I got a bit confused on the conclusions of these steps. Because the Sum of weights seems to say that the forest type has an influence on density. However, when using the expand.grid() function for predicting for multiple covariates, shows that none of the covariates has an effect. Not sure I am interpreting this right…
Below the code and results I got.
CI_dens
0.025 0.975
lam(Int) -0.1427598 0.1986864
lam(ForestAnkarafa) 0.8081677 1.1855487
CIDetect
0.025 0.975
p(Int) 1.8330838 2.122539
p(CorePercent) 0.4815181 1.094935
predictionsFO <- cbind(
ForestDF,
predict(
hnCO_FO,
newdata = ForestDF,
type="state"))
predictionsFO
Forest Predicted SE lower upper
1 Anabohazo 1.028358 0.08957533 0.8669623 1.219799
2 Ankarafa 2.786598 0.18336074 2.4494268 3.170182
core_percent <- data.frame(
CorePercent = c(0, 0.5, 1))
predictionsCO <- cbind(core_percent,
predict(hnCO_FO,
newdata = core_percent,
type="det"))
predictionsCO
CorePercent Predicted SE lower upper
1 0.0 7.226908 0.5336490 6.253140 8.352316
2 0.5 10.718001 0.4925153 9.794883 11.728119
3 1.0 15.895531 1.6682455 12.940198 19.525814
PredCO_FO <- cbind(cov_combinations,
round(
predict(hnCO_FO,
newdata = cov_combinations,
type="state")
, 3))
PredCO_FO
CorePercent Forest Predicted SE lower upper
1 0.00 Anabohazo 1.028 0.090 0.867 1.22
2 0.25 Anabohazo 1.028 0.090 0.867 1.22
3 0.50 Anabohazo 1.028 0.090 0.867 1.22
4 0.75 Anabohazo 1.028 0.090 0.867 1.22
5 1.00 Anabohazo 1.028 0.090 0.867 1.22
6 0.00 Ankarafa 2.787 0.183 2.449 3.17
7 0.25 Ankarafa 2.787 0.183 2.449 3.17
8 0.50 Ankarafa 2.787 0.183 2.449 3.17
9 0.75 Ankarafa 2.787 0.183 2.449 3.17
10 1.00 Ankarafa 2.787 0.183 2.449 3.17

 to your question
to your question to support your conclusion
to support your conclusion with confidence intervals
with confidence intervals on your next step, challenge you solved, problem you still have or leap of understanding
on your next step, challenge you solved, problem you still have or leap of understanding to a coursemate
to a coursemate