Hi all, here is my second assignment.
1) Plots:
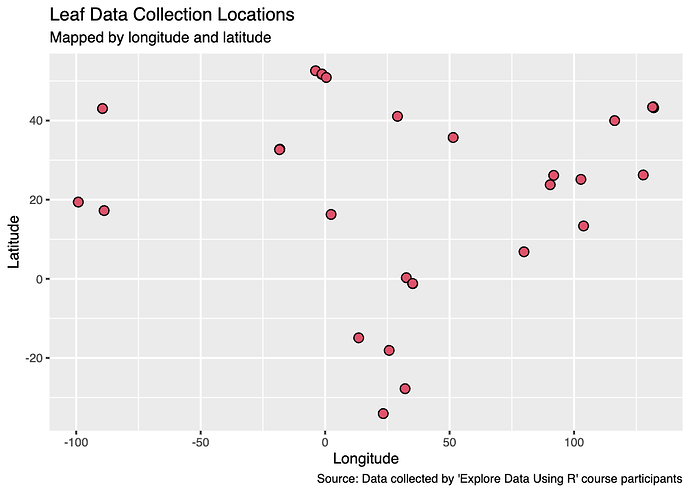
a) Using a scatterplot for more of an anticipatory applied use, I plotted Longitude and Latitude of the leaf data, to effectively create a map of collection locations. I customised the plot points and labels.
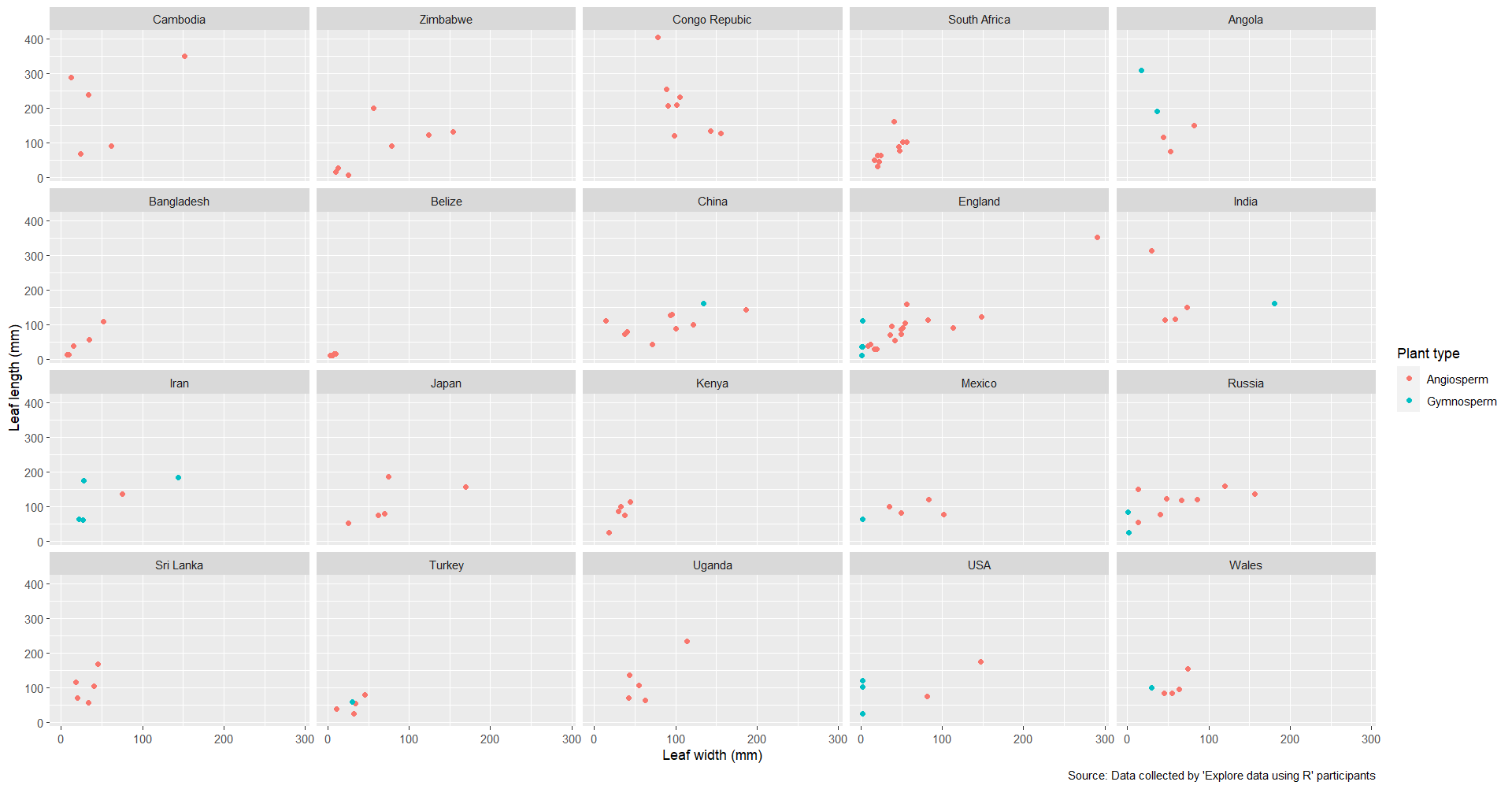
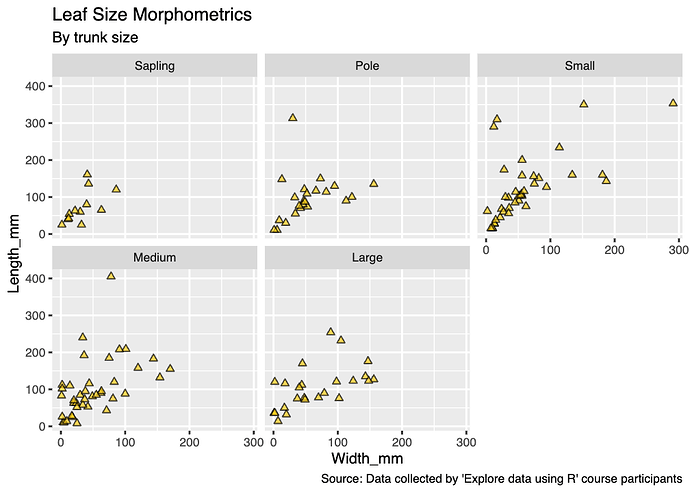
b) I also made a multi-plot figure of leaf morphometrics (width and length) by trunk
size, again customising the plot points and text.
2) Code:
a)
ggplot(LeafData,
aes(x = Longitude, y = Latitude)) +
geom_point(size = 3, shape = 21, colour = 367777, fill = 336666) +
labs(title = "Leaf Data Collection Locations",
subtitle = "Mapped by longitude and latitude",
caption = "Source: Data collected by 'Explore data using R' course participants")
b)
ggplot(LeafData,
aes(x = Width_mm, y = Length_mm)) +
geom_point(size = 1.7, shape = 24, colour = 367777, fill = 399183, alpha = 0.67) +
facet_wrap(vars(TrunkSize)) +
labs(title = "Leaf Size Morphometrics",
subtitle = "By trunk size",
caption = "Source: Data collected by 'Explore data using R' course participants")
3) Interpretation:
a) Though there cannot be assumptions made like with a traditional scatterplot, it does show that most leaf data was collected from locations in the global north.
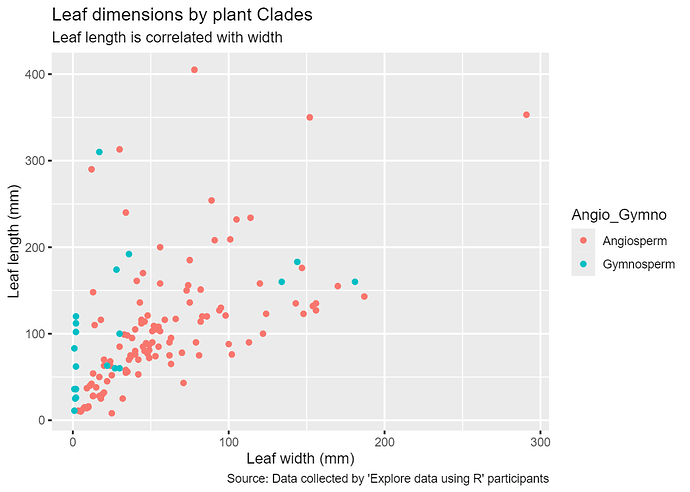
b) It appears that a positive correlation between leaf length and width is maintained across trunk sizes.
4) Brief reflection:
I’m looking forward to tweaking the code to try this all out on different plot types and with my own data, as well as exploring additional plot customisations and colour scheme options.
5) Feedback:
@Rachel_Crouthers, beautiful & complex multi-plot figure! I also have similar questions to the ones you posed in your assignment.
And, regarding my general comment on the first assignment, it was great to find that scale_colour_viridis_d seems to be the best option for colourblindness!
 that you’ve created during this module
that you’ve created during this module the plot; what meaning do you draw?
the plot; what meaning do you draw? next step, challenge you solved, or problem you still have
next step, challenge you solved, or problem you still have to a coursemate
to a coursemate